In classification problems, the target variable can be one of a discrete set of categories or classes and the goal is to predict the target based on a set of inputs or predictor variables. Tackling a classification problem is an example of supervised machine learning. In this article, we take a look at one algorithm for classification: support vector machines.
Table of Contents
What Are Support Vector Machines?
Support Vector Machine (SVM) algorithms for classification attempt to find boundaries that separate the different classes of the target variables. The boundaries are found by maximizing the distance between points closest to the boundaries on either side. These data points are the “support vectors” that we focus on to determine how to classify the rest of the data.
For example, SVMs can be used to predict cancer occurrence (Present, Not Present) based on different physiological characteristics, or college academic performance based on high school GPA and standardized test scores. SVM is a very robust algorithm that can be used for a wide variety of applications.
Linear and Non-Linear SVM
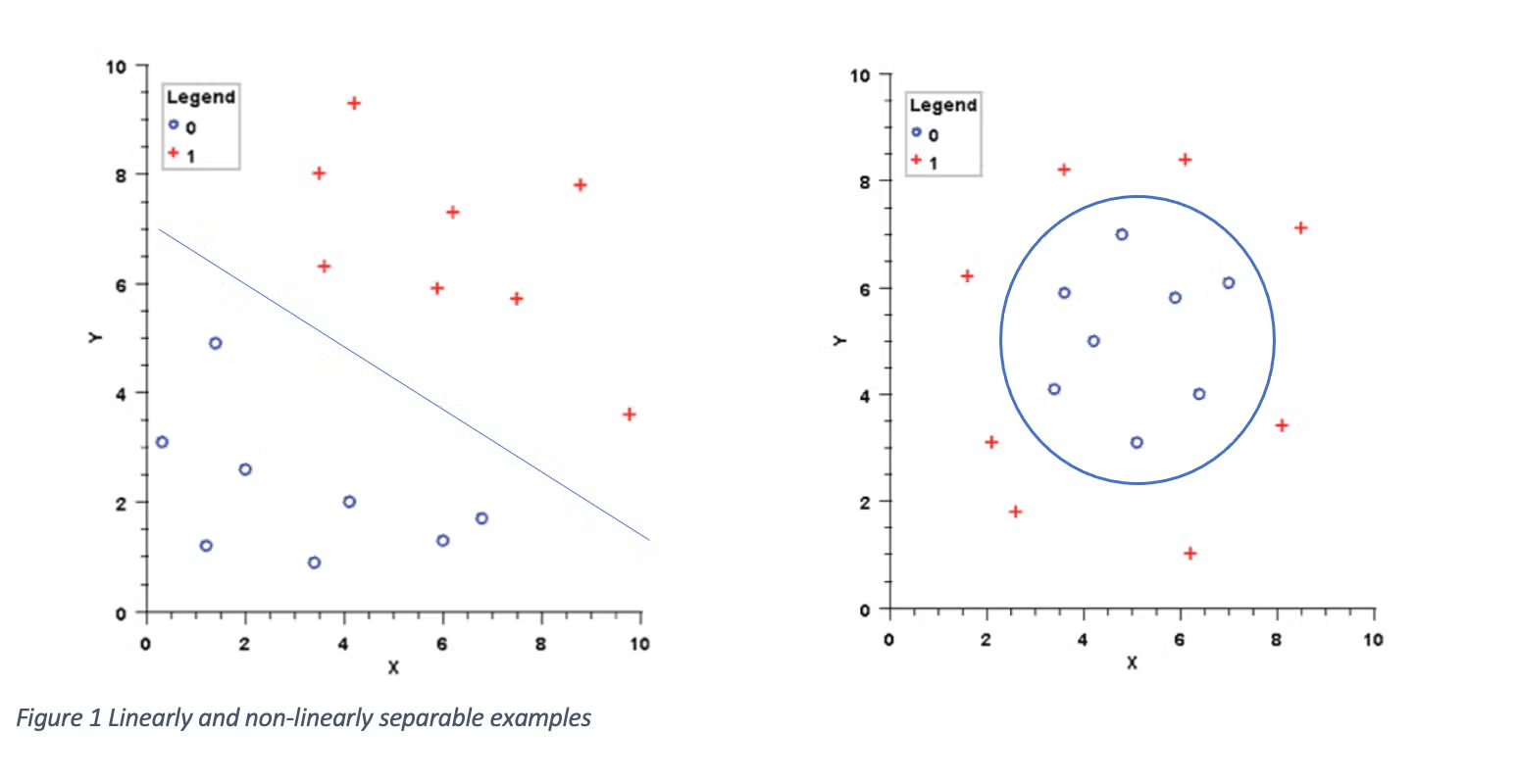
Below is a graphical representation of how an SVM works for a target variable with two classes. The line represents the dividing line that maximizes the distance between the line and the closest points from each group. Once the dividing line is defined, we can classify new points by finding which side of the line they fall on.
Situations when the classifications can be separated with a line are called linearly-separable. In more complex (higher dimensions) cases, the dividing line becomes a plane or hyperplane dividing the groups. You may also see some data sets where the groups are divided in a non-linear pattern.
 Back to top
Back to top
SVM and Machine Learning
Support vector machines fit into the data science process the same way a regression model or any other predictive model does. Given a dataset containing both the inputs and the target variable, a model is trained on a (randomly selected) subset of the data. Then the trained model is used to predict outcomes on a separate validation set. By repeating the process of randomly partitioning the dataset into a training set and validation set, different configurations of the model can be compared. This is sometimes called model tuning because it helps fine-tune model settings. For support vector machines, an important configuration parameter is the choice of a kernel function.
SVM Kernel Trick
Through the SVM kernel trick, the original points are mapped from their original configuration (the input space) into a modified destination (the feature space), where they become linearly-separable. There are different kinds of kernel functions and selecting an appropriate kernel function is part of configuring an SVM model suitable to a specific problem. The math for this is complicated, but IMSL makes this part of the process easier by providing easy settings for different kernels.
Back to topSVM and IMSL
With IMSL, you can follow the general process of applying an SVM to a dataset and evaluating its performance. Here is an example using the Statlog (Heart) Data Set from the UCI Machine Learning Repository. The data contains various characteristics and lab test results on 270 patients with and without heart disease. The goal is to train a model to predict (or diagnose) the absence or presence of heart disease based on these predictors:
- age
- sex
- chest pain types (4 values)
- resting blood pressure
- serum cholesterol in mg/dl
- fasting blood sugar 120 mg/dl
- resting electrocardiographic results (values 0,1,2)
- maximum heart rate achieved
- exercise induced angina
- oldpeak = ST depression induced by exercise relative to rest
- the slope of the peak exercise ST segment
- number of major vessels (0-3) colored by fluoroscopy
- thal: 3 = normal; 6 = fixed defect; 7 = reversible defect
Using SVM algorithms in IMSL for Java follows these basic steps:
// Define the input data types:
SVClassification.VariableType[] varType = {
SVClassification.VariableType.QUANTITATIVE_CONTINUOUS, //age
SVClassification.VariableType.CATEGORICAL, //sex
SVClassification.VariableType.CATEGORICAL, //chest pain type
SVClassification.VariableType.QUANTITATIVE_CONTINUOUS, //blood pressure
SVClassification.VariableType.QUANTITATIVE_CONTINUOUS, // cholesterol
SVClassification.VariableType.QUANTITATIVE_CONTINUOUS, //blood sugar
SVClassification.VariableType.CATEGORICAL, //ecg
SVClassification.VariableType.QUANTITATIVE_CONTINUOUS, //max heart rate
SVClassification.VariableType.QUANTITATIVE_CONTINUOUS, //induced angina
SVClassification.VariableType.QUANTITATIVE_CONTINUOUS, //ST depression
SVClassification.VariableType.ORDERED_DISCRETE, //slope
SVClassification.VariableType.QUANTITATIVE_CONTINUOUS, //number of vessels
SVClassification.VariableType.CATEGORICAL, //thal
SVClassification.VariableType.CATEGORICAL, //heart disease(absent/present)
};
final int N = 270; // total number of observations
final int N_TEST = 25; // number of observations to use for testing
final int N_TRAIN = N - N_TEST; // number of observations to use for training
final int N_COLS = 14; // number of columns in the data (count)
final int TARGET_COl = 13; // target variable column (0-based)
// Reads the data from a file and formats it as an array
double[][] x_full = getData() ;
// Randomly select a training set and a test (validation) set
RandomSamples rs = new RandomSamples(new Random(134567));
int[] idx = rs.getPermutation(N);
double[][] x_train = new double[N_TRAIN][N_COLS];
double[][] x_test = new double[N_TEST][N_COLS];
// Populate the training data and test (validation) set
for (int i = 0; i < N_TEST; i++) {
x_test[i] = x_full[idx[i] - 1];
}
for (int i = 0; i < N_TRAIN; i++) {
x_train[i] = x_full[idx[i+N_TEST] - 1];
}
// Construct a Support Vector Machine and train it using training data
SVClassification svm = new SVClassification(x_train, TARGET_COl, varType);
// Use the linear kernel
svm.setKernel(new LinearKernel());
// Train the model
svm.fitModel();
//Predict an outcome on the test data and compare to the known outcomes:
double[] predictedClass = svm.predict(x_test);
int[][] predictedClassErrors =
svm.getClassErrors(knownClassTest, predictedClass);
The following confusion matrix shows the results of the prediction:
| Predicted | Predicted | |
|---|---|---|
| Actual | Absent | Present |
| Absent | 12 | 2 |
| Present | 2 | 9 |
For this validation set, our SVM predicts heart disease in two out of 14 who do not have heart disease. In two out of 11 with heart disease, the model predicts that they don’t have heart disease. Overall 21/25 are diagnosed correctly in this validation set. The second type of error (incorrectly predicting the absence of heart disease when there is heart disease) is known as a false negative and is usually considered a more severe type of error. We could tune the model further to attempt to minimize overall errors, but especially the false negatives.
Back to topConclusion
Support vector machines can be a robust tool for classification problems complementing other data mining techniques. The IMSL for Java library implements a flexible programming interface for users to explore this modern technique as shown in the examples in this article.
Want to try JMSL on in your application? Request a free trial.
Note: This post was originally published on September 19, 2016 and has been updated for accuracy and comprehensiveness.
